-
PDF
- Split View
-
Views
-
Cite
Cite
Yick Fu Wong, Tak Hong Cheung, Keith Wing Kit Lo, So Fan Yim, Loucia Kit Yin Chan, Olivier Buhard, Alex Duval, Tony Kwok Hung Chung, Richard Hamelin, Detection of microsatellite instability in endometrial cancer: advantages of a panel of five mononucleotide repeats over the National Cancer Institute panel of markers, Carcinogenesis, Volume 27, Issue 5, May 2006, Pages 951–955, https://doi.org/10.1093/carcin/bgi333
Close - Share Icon Share
Abstract
The aim of this study was to find the optimal set of microsatellite markers for diagnosis of the microsatellite instability (MSI) phenotype in endometrial cancers. We compared the sensitivity, specificity and ease of use of a reference panel of five markers originally recommended by the National Cancer Institute (NCI) for colorectal cancer and a panel of five quasi-monomorphic mononucleotide repeat markers (pentaplex PCR system). We used these panels for establishing the MSI status of a series of 80 sporadic endometrial adenocarcinomas by comparing the allelic profiles of the markers between tumor and matching germline DNA. Both panels detected the same subset of 21 out of 80 (26%) endometrial MSI carcinomas. However, in the MSI cases, the mean instability of the five mononucleotide repeats was 96.1% as compared with a mean instability of 69.8% for the three dinucleotide repeats of the NCI panel, indicating a superiority of mononucleotide repeats over dinucleotide repeats in detecting MSI. The fact that the two panels of markers detect the same set of MSI tumors is due to the presence of two mononucleotide repeats within the NCI panel. As demonstrated previously in gastric and colon MSI cases, the pentaplex PCR reaction using mononucleotide repeats is thus an easier and more sensitive method than the NCI panel, for the screening of MSI status in endometrial tumors.
Introduction
Endometrial cancer is a common malignant tumor in women worldwide. Its incidence was 10.4 per 100 000 in Hong Kong women in 1998. Although it has a relatively low mortality rate, some behave aggressively. Most endometrial cancers are sporadic and these comprise different histological subtypes. Approximately 80% are endometrioid type and 10% are serous. These two types of endometrial adenocarcinoma have different prevalence, etiology and prognosis and may also involve different genetic events in their pathogenesis and progression.
Microsatellite instability (MSI) is defined by the presence of alternate-sized repetitive sequences in tumor DNA that is not seen in the corresponding germline DNA ( 1 – 3 ). It is recognized as the hallmark of a defect in the mismatch repair system ( 4 – 7 ). Originally, MSI was described in hereditary non-polyposis colorectal cancer (HNPCC) as well as in sporadic colorectal tumors ( 8 ). The ‘International Workshop on Microsatellite Instability and RER Phenotypes in Cancer Detection and Familial Predisposition’, sponsored by the National Cancer Institute (NCI), recommended a panel of five markers (two mononucleotide markers BAT-26 and BAT-25 and three dinucleotide markers D5S346, D2S123 and D17S250) for the uniform analysis of MSI in colorectal tumors ( 8 ). Tumors with instability at two or more of these markers were defined as being high-frequency MSI (MSI-H), whereas those with instability at one repeat or showing no instability were defined as low-frequency MSI (MSI-L) or microsatellite stable (MSS) tumors, respectively ( 8 ). The use of dinucleotide repeat microsatellite markers has, however, been shown to lead to confusion in the screening of gastrointestinal cancers for MSI ( 9 ), and mononucleotide repeats were shown to be more specific and sensitive ( 9 – 11 ). Recently, a pentaplex PCR system composed of five quasi-monomorphic mononucleotide repeats (BAT-25, BAT-26, NR-21, NR-22 and NR-24) was described to establish tumor MSI status with 100% specificity and sensitivity and without the need for evaluating corresponding germline DNA ( 12 ).
MSI has also been reported in endometrial carcinoma ( 13 – 16 ), but the optimal set of loci for diagnosing MSI in these tumors may well be different. The aim of the present study was to compare the NCI panel and the pentaplex system for the detection of MSI in a series of endometrial tumors. This was performed by analysing PCR-amplified profiles of each marker in tumor and matching normal DNA. The possibility for detection of endometrial MSI tumors using the pentaplex without reference to matched normal DNA was also examined. This study validates a method for the detection of MSI in endometrial cancer, thus allowing better classification of this tumor type and subsequent improvement in prognostic information for patient management.
Materials and methods
Tumor samples
Eighty invasive endometrioid endometrial carcinoma and matching blood samples were used. This study was approved by the Clinical Research Ethics Committee of the Chinese University of Hong Kong, and tissues were collected from study subjects after obtaining informed consent. Both tissue and blood samples were snap-frozen in liquid nitrogen and stored at −80°C. Tissue samples were embedded in OCT compound until required for analysis. Consecutive thin sections of frozen tissue were prepared by cryomicrotome, with the first and final sections stained with H & E and used for guiding the removal of malignant cells. Enriched tumor cells were recovered from a minimum of 10 consecutive sections and transferred into sterile microtubes moistened with lysis buffer. DNA from neoplastic cells was extracted using PURIGENE ® DNA Isolation Kits (Gentra Systems, Minneapolis, MN, USA) according to manufacturer's instructions. DNA from blood cells was prepared by proteinase K digestion and phenol/chloroform extraction.
Microsatellite analysis
The five markers in pentaplex panel 1 were mononucleotide repeats BAT-25, BAT-26, NR-21, NR-24 and NR-27. Primer sequences were as described previously ( 17 ), and each sense primer was end-labelled with one of the fluorescent markers FAM, HEX or NED. Pentaplex PCR was performed in a Perkin-Elmer 9600 thermocyclerŠ (Norwalk, CT) with an initial 5 min denaturation step at 94°C, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 7 min. Amplified PCR products were run on an Applied Biosystems PRISM 310 automated capillary electrophoresis DNA sequencer. Allelic sizes were estimated using Genescan 2.1 software (Applied Biosystems, Foster City, LA, USA).
The markers in panel 2 were those recommended by NCI and included three dinucleotide markers D2S123, D5S346, and D17S250 and the two mononucleotide repeats BAT-25 and BAT-26 ( 8 ). For dinucleotide markers, single PCR were carried out with sense primers end-labelled with fluorescent markers FAM, HEX or NED, respectively. Amplification was started with 12 min at 95°C, followed by 10 cycles composed of 15 s at 94°C, 15 s at 55°C and 30 s at 72°C. This was followed by 25 cycles composed of 15 s at 89°C, 15 s at 55°C and 30 s at 72°C. Amplified PCR products for the three dinucleotide markers were pooled and run on an ABI PRISM 310 DNA sequencer. Data for panel 2 were then complemented with results obtained for BAT-25 and BAT-26 in the pentaplex reaction in order to obtain the final NCI panel information. To exclude the possibility of technical artifacts or contamination, all observed differences were reproduced by independent determination.
Results
Reading of dinucleotide repeat profiles
Allelic profiles for the D2S123, D5S346 and D17S230 dinucleotide repeats were individually compared in the series of 80 endometrial cancer tissues against matched blood germline DNA samples. These markers are highly polymorphic and, hence, show different profiles between individuals. In the majority of cases, they show reproducible profiles, with tumor DNA and matched normal DNA showing superimposable results. On some occasions one of the alleles was not amplified or amplified with lower intensity in tumor DNA as compared with germline DNA, indicating loss of heterozygosity (LOH). A positive result for instability was scored when amplified alleles from tumor DNA were not present in amplified normal DNA, even when changes were minor. D2S123, D5S346 and D17S250 showed instability in 16, 13 and 17 of the 80 analyzed tumor samples, respectively (light grey cells in Table I ).
MSI-H and MSI-L endometrial tumors
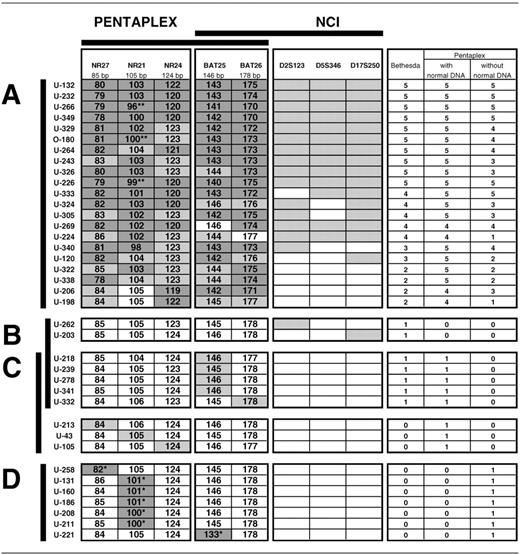
MSI-H tumors detected with the NCI panel of markers or with the pentaplex with reference to matching normal DNA ( A ). MSI-L tumors detected with the NCI panel ( B ) and the pentaplex with ( C ) or without ( D ) reference to matching normal DNA.
Dark grey cells are unstable alleles of mononucleotide repeats outside of QMVR of each marker that could be detected without reference to matching normal DNA, while light grey cells are unstable alleles detectable only when compared with normal DNA. Polymorphisms are indicated by asterisks, ( ** ) when showing additional instability in tumor DNA and ( * ) when not.
MSI-H and MSI-L endometrial tumors

MSI-H tumors detected with the NCI panel of markers or with the pentaplex with reference to matching normal DNA ( A ). MSI-L tumors detected with the NCI panel ( B ) and the pentaplex with ( C ) or without ( D ) reference to matching normal DNA.
Dark grey cells are unstable alleles of mononucleotide repeats outside of QMVR of each marker that could be detected without reference to matching normal DNA, while light grey cells are unstable alleles detectable only when compared with normal DNA. Polymorphisms are indicated by asterisks, ( ** ) when showing additional instability in tumor DNA and ( * ) when not.
Reading of mononucleotide repeat profiles
Two different readings were performed with the mononucleotide repeats NR21, NR24, NR27, BAT-25 and BAT-26, for each of the 80 samples. First, amplification profiles with tumor DNA were compared with amplification profiles obtained from matching normal DNA. As in the case of dinucleotide repeats, allelic profiles obtained from tumor DNA and matched normal DNA were generally almost perfectly superimposable. Any changes in tumor DNA as compared with matched normal DNA, even if minor, were considered to represent instability (dark and light grey cells in Table I ).
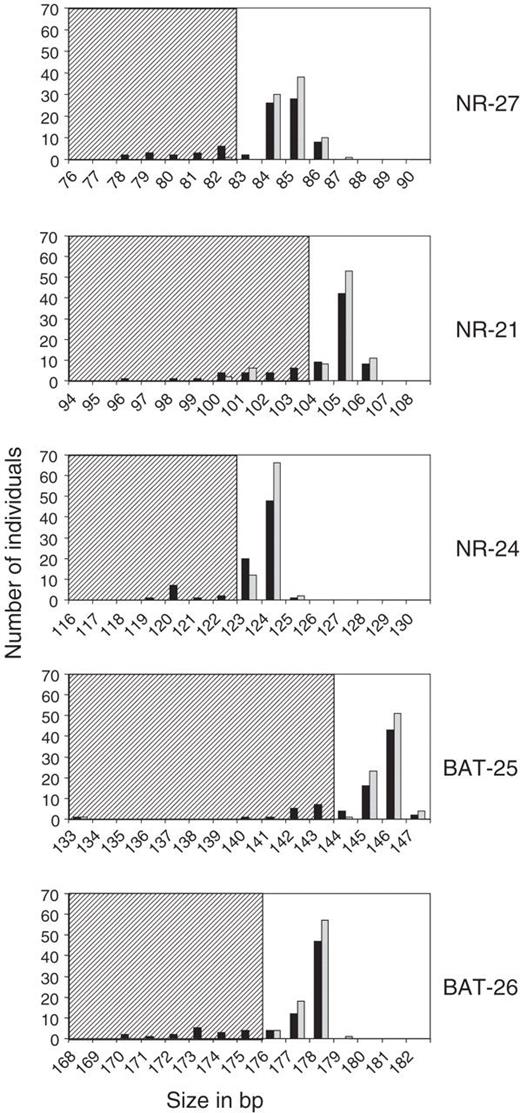
For each marker, Genescan 2.1 software was used for calculating the size of PCR products compared with the size of internal standards. We measured the sizes of the shortest alleles for each marker amplified from both normal and tumor DNA from the 80 samples ( Figure 1 ). Alleles obtained from germline DNA define a quasi-monomorphic variation range (QMVR) for each marker. A few cases showed distinct sizes outside the QMVR and represent rare variant alleles. Allelic sizes within the QMVR are frequent in the complete series of endometrial tumor DNA. Markers showing instability as compared with matching normal DNA, are in most cases, but not all, in the striped zones outside the QMVR ( Figure 1 ). The sizes of the shortest alleles in tumors showing instability for any of the markers are also indicated in Table I . Samples U-266, O-180 and U-226 present a polymorphism for NR-21 in normal DNA and instability of this variant allele in tumor DNA (indicated by ** in Table I ). Samples U-258 for NR-27, U-131, U-160, U-186, U-208 and U-211 for NR-21 and U-221 for BAT-25 present a polymorphism in the normal DNA with no additional size variation in tumor DNA (indicated by * in Table I ). Therefore, NR-27, NR-21, NR-24, BAT-25 and BAT-26 showed instability in 22, 20, 22, 24 and 21 of the 80 tumor samples, respectively, when compared against matching normal DNA (light and dark grey cells in Table I , with the exception of * cells).
Number of individuals with the indicated allelic sizes in tumor (black) and germline (grey) DNA for each marker of the pentaplex. Striped zones are outside the QMVR and contain alleles either showing instability in tumor DNA (without reference to matched normal DNA), or polymorphism in germline DNA.
The PCR amplification profiles of tumor DNA were then read again without consideration of the matched normal DNA profiles. Each marker with a size outside of the QMVR was considered unstable. Because normal DNA was not examined, every sample with a polymorphism in any of the mononucleotide repeats was thus scored positive for instability. In these circumstances, NR-27, NR-21, NR-24, BAT-25 and BAT-26 showed instability in 16, 21, 11, 15 and 17 out of the 80 tumor samples, respectively (dark grey cells in Table I ). The light grey cells correspond to markers showing instability in tumor samples when compared with matched normal DNA, but where size variation was not important enough to be detected without such a comparison because allelic sizes remained within the QMVR.
MSI determined by the NCI panel of markers
According to the consensus meeting guidelines that require instability in at least 2 of the 5 markers ( 8 ), 21 out of 80 tumors (26%) were MSI-H ( Table I , subset A). These tumors show instability at 5 (10 cases), 4 (5 cases), 3 (2 cases) and 2 markers (4 cases) ( Table I ). Taken independently, BAT-25, BAT-26, D2S123, D5S346 and D17S250 show instability in 20 (95%), 20 (95%), 15 (71%), 13 (62%) and 16 (76%) of the 21 MSI-H tumors, respectively.
A tumor is defined as MSI-L if it shows instability at only one of the five markers ( 8 ). Accordingly, seven tumors were MSI-L ( Table I , subset B) because they showed instability only at BAT-25 (4 cases), BAT-26 (1 case), D2S123 (1 case) or D17S250 (1 case). The remaining 52 tumors were classified MSS with the NCI panel of markers.
MSI determined by the pentaplex system with matched normal DNA
If we use the pentaplex system of five mononucleotide repeats for comparing allelic profiles between normal and tumor DNA, then 21 of the 80 analyzed cases would be classified as MSI-H tumors according to the consensus meeting guidelines requiring a minimum of two repeats showing instability. These cases were identical to those detected with the NCI panel, but they showed instability at 5 (17 cases) or 4 (4 cases) markers ( Table I , subset A). Individually, NR-27, NR-21, NR-24, BAT-25 and BAT-26 show instability in 21 (100%), 19 (90%), 21 (100%), 20 (95%) and 20 (95%) of the 21 MSI-H tumors, respectively.
Eight tumors can be considered as MSI-L since they showed instability only at NR-27 (1 case), NR-21 (1 case), NR-24 (1 case), BAT-25 (4 cases) or BAT-26 (1 case) ( Table I , subset C). It has to be noted that these particular amplification profiles could rather be due to a LOH in the corresponding loci in tumor DNA as compared with matching normal DNA with two alleles with 1 or 2 bp size differences. But to keep our reading more stringent and more strict against mononucleotide repeats, we considered them as unstable. The five cases showing instability with BAT-25 or BAT-26 are common to MSI-L tumors as defined by the NCI panel. The remaining 51 tumors are classified MSS with this pentaplex panel using matched normal DNA.
MSI determined by the pentaplex system without matched normal DNA
For each mononucleotide repeat, only dark grey cells in Table I are considered in this reading since they represent markers with sizes outside the QMVR. Owing to the known presence of ethnic-related polymorphisms in these markers, and in the absence of matching normal DNA, we have previously proposed that a tumor should be considered MSI-H when it shows instability for at least three of the five markers of the pentaplex panel ( 12 ). According to this rule, 16 of the 21 MSI-H tumors defined with the NCI and/or the pentaplex panels with matched normal DNA are detected with the pentaplex system in the absence of normal DNA.
Of the five remaining identified MSI-H samples, cases U-120, U-322 and U-338 show clear instability on two markers and cases U-224 and U-198 on only one marker, and therefore are not considered MSI-H in this reading. Among the 59 other tumors, 7 cases show clear shortening of one of the mononucleotide repeats (with an * in Table I , subset D). Because of the earlier readings, we know that these markers present polymorphisms, but this fact cannot be deduced without analysis of matching normal DNA. It has, however, no consequence on the reading since these samples are considered as non-MSI tumors.
Discussion
The NCI panel of microsatellite markers was proposed at a consensus meeting to determine MSI tumors in colorectal cancers, and in the absence of specific markers, in other localizations ( 8 ). We recently showed that a pentaplex PCR system composed of five quasi-monomorphic mononucleotide repeats was easier to use and more sensitive and specific than the NCI panel of markers for determining MSI status ( 12 , 17 ). This pentaplex system is recommended in Table I of the report of the most recent NCI consensus meeting on MSI and HNPCC tumors ( 18 ).
By comparing the amplification profiles of tumor DNA with matched normal DNA in a series of 80 endometrioid-type endometrial carcinomas, we have shown here that the NCI panel of markers and the pentaplex system of mononucleotide repeats resulted in detection of the same 21 MSI-H tumors. It appears, however, that the two methods have different properties. The NCI panel revealed instability in 84 out of 105 (77%) markers in the MSI-H tumors compared with instability in 101 out of 105 (96%) markers of the pentaplex panel. When comparison is made between dinucleotide and mononucleotide repeats, the difference is even more apparent and is statistically different. Dinucleotide repeats have a sensitivity of 44 out of 63 (70%) compared with 101 out of 105 (96%) for the mononucleotide repeats ( P = 0.0001). For both methods, we used the same cut-off value of two markers of five showing instability in order to define a tumor as MSI-H. MSI-H tumors showed instability at 5 (10 cases), 4 (5 cases), 3 (2 cases) and 2 markers (4 cases) with the NCI panel, while they showed instability at 5 (17 cases) or 4 (4 cases) of the 5 mononucleotide repeats in the pentaplex system. Moreover, four MSI-H tumors (U322, U338, U206 and U198) were unstable only at the two monucleotide repeats (BAT-25 and BAT-26) of the NCI panel, with no instability in the dinucleotide repeats. The same tumors were unstable in 5 (2 cases) or 4 (2 cases) of the markers of the pentaplex. We can underline that the analysis of BAT-26 and BAT-25 alone would have been sufficient for detecting MSI in almost all tumors since both these markers were unstable, when compared with matching normal DNA, in 19 out of 21 of the MSI-H tumors, the two remaining tumors (U224 and U269) showing instability on only one of these repeats. All these observations indicate that mononucleotide repeats are much more unstable in MSI-H endometrial tumors than dinucleotide repeats, as it was already observed in MSI-H gastrointestinal tumors ( 9 , 10 , 12 ).
We have shown previously that owing to the quasi-monomorphic nature of the mononucleotide repeats in Caucasians, the pentaplex system could be used for determining the MSI status in gastrointestinal tumors of this population, without reference to matching normal DNA ( 12 ). This observation was recently extended to a worldwide multipopulation analysis ( 19 ). In the present population of Hong Kong Chinese, we showed that polymorphisms in these markers were infrequent. The only exception was NR-21 with an allelic variant in 8 out of 80 (10%) of the individuals. Therefore, without reference to normal matched DNA, no false-positive MSI-H tumors would have been determined. Using the rule of at least three markers showing instability for a tumor to be defined as MSI-H, we would have detected 16 out of the 21 MSI-H tumors. Finally, five samples would have been negative (U224, U120, U322, U338 and U198) and therefore misclassified. This is not surprising since it is already known that MSI-H endometrial tumors show a lower intensity of instability compared with gastrointestinal MSI-H tumors at both coding and non-coding repeats ( 20 , 21 ). In contrast to gastrointestinal tumors, analysis of matched normal DNA analysis is thus required for accurate MSI-H evaluation in endometrial cancer with the pentaplex.
Taken together, our results indicate that the best markers within the NCI panel are the mononucleotide repeats BAT-25 and BAT-26. Other mononucleotide repeats of the pentaplex are as effective as BAT-25 and BAT-26. In the present endometrial cancer series, NR27, NR24, BAT-25, BAT-26, NR-21, D17S250, D2S123 and D5S346 showed instability in 100, 100, 95, 95, 90.5, 76, 71.5 and 62% of the 21 MSI-H tumors, respectively. We believe that the pentaplex system of mononucleotide repeats is superior in terms of accuracy and ease of use compared with the NCI panel of markers for determination of the MSI status of endometrial cancers. Similar comparisons should be performed for other tumor types, next to gastrointestinal tumors where it is already done, that potentially harbor the MSI-H phenotype.
This study was supported by Hong Kong–France Joint Research Scheme (Procore Egide) F-HK07/03T. The authors would like to thank the Gynaecologic Cancer Research Laboratory of the Chinese University of Hong Kong in providing equipment. We thank Dr Barry Iacopetta for critical reading of the manuscript.
Conflict of Interest Statement: None declared.
References
Ionov,Y., Peinado,M., Malkhosyan,S., Shibata,D. and Perucho,M. (
Aaltonen,L.A., Peltomäki,P., Leach,F.S. et al . (
Thibodeau,S.N., Bren,G. and Schaid,D. (
Bronner,C.E., Baker,S.M., Morrison,P.T. et al . (
Papadopoulos,N., Nicolaïdes,N.C., Wei,Y.F. et al . (
Fishel,R., Lescoe,M.K., Rao,M.R.S., Copeland,N.G., Jenkins,N.A., Garber,J., Kane,M. and Kolodner,R. (
Leach,F.S., Nicolaïdes,N.C., Papadopoulos,N. et al . (
Boland,C.R., Thibodeau,S.N., Hamilton,S.R. et al . (
Perucho,M. (
Hoang,J.M., Cottu,P.H., Thuille,B., Salmon,R.J., Thomas,G. and Hamelin,R. (
Zhou,X.P., Hoang,J.M., Li,Y.J. et al . (
Suraweera,N., Duval,A., Reperant,M., Vaury,C., Furlan,D., Leroy,K., Seruca,R., Iacopetta,B. and Hamelin,R. (
Risinger,J.L., Berchuck,A., Kohler,M.F., Watson,P., Lynch,H.T. and Boyd,J. (
Kobayashi,K., Sagae,S., Kudo,R., Saito,H., Koi,S. and Nakamura,Y. (
Helland,A., Borresen-Dale,A.L., Peltomaki,P., Hektoen,M., Kristensen,G.B., Nesland,J.M., de la Chapelle,A. and Lothe,R.A. (
Simpkins,S.B., Bocker,T., Swisher,E.M., Mutch,D.G., Gersell,D.J., Kovatich,A.J., Palazzo,J.P., Fishel,R. and Goodfellow,P.J. (
Buhard,O., Suraweera,N., Lectard,A., Duval,A. and Hamelin,R. (
Umar,A., Boland,C.R., Terdiman,J.P. et al . (
Buhard,O., Cattaneo,F., Wong,Y.F., Yim,S.F., Friedman,E., Flejou,J.F., Duval,A. and Hamelin,R. (
Schwartz,S., Yamamoto,H., Navarro,M., Maestro,M., Reventos,J. and Perucho,M. (
Author notes
Department of Obstetrics and Gynaecology, The Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, N.T., Hong Kong, China and 1Inserm, U762, Paris, F-75010 France;, Univ Paris 6, Paris, F-75005 France