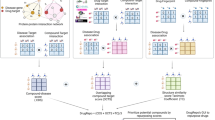
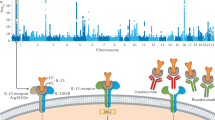
Abstract
Given the high attrition rates, substantial costs and slow pace of new drug discovery and development, repurposing of 'old' drugs to treat both common and rare diseases is increasingly becoming an attractive proposition because it involves the use of de-risked compounds, with potentially lower overall development costs and shorter development timelines. Various data-driven and experimental approaches have been suggested for the identification of repurposable drug candidates; however, there are also major technological and regulatory challenges that need to be addressed. In this Review, we present approaches used for drug repurposing (also known as drug repositioning), discuss the challenges faced by the repurposing community and recommend innovative ways by which these challenges could be addressed to help realize the full potential of drug repurposing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Ashburn, T. T. & Thor, K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 (2004).
Scannell, J. W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Pammolli, F., Magazzini, L. & Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 10, 428–438 (2011).
Waring, M. J. et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14, 475–486 (2015).
Roundtable on Translating Genomic-Based Research for Health (Board on Health Sciences Policy) Institute of Medicine in Drug Repurposing and Repositioning: Workshop Summary (eds. Johnson, S. G., Beachy, S. H., Olson, S., Berger, A. C.) (National Academies Press, Washington DC, 2014).
Breckenridge, A. & Jacob, R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. https://doi.org/10.1038/nrd.2018.92 (2018).
Nosengo, N. Can you teach old drugs new tricks? Nature 534, 314–316 (2016).
Phillips, D. J. Pfizer's expiring Viagra patent adversely affects other drugmakers too. Forbes https://www.forbes.com/sites/investor/2013/12/20/pfizers-expiring-viagra-patent-adversely-affects-other-drugmakers-too (2013).
Singhal, S. et al. Antitumor activity of thalidomide in refractory multiple myeloma. N. Engl. J. Med. 341, 1565–1571 (1999).
Urquhart, L. Market watch: top drugs and companies by sales in 2017. Nat. Rev. Drug Discov. 17, 232 (2018).
Hurle, M. R. et al. Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther. 93, 335–341 (2013).
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Hieronymus, H. et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330 (2006).
Dudley, J. T., Deshpande, T. & Butte, A. J. Exploiting drug-disease relationships for computational drug repositioning. Brief Bioinform. 12, 303–311 (2011).
Iorio, F., Rittman, T., Ge, H., Menden, M. & Saez-Rodriguez, J. Transcriptional data: a new gateway to drug repositioning? Drug Discov. Today 18, 350–357 (2013).
Dudley, J. T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl Med. 3, 96ra76 (2011).
Sirota, M. et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl Med. 3, 96ra77 (2011).
Wagner, A. et al. Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol. Syst. Biol. 11, 791 (2015).
Hsieh, Y. Y., Chou, C. J., Lo, H. L. & Yang, P. M. Repositioning of a cyclin-dependent kinase inhibitor GW8510 as a ribonucleotide reductase M2 inhibitor to treat human colorectal cancer. Cell Death Discov. 2, 16027 (2016).
Huang, C. H. et al. Identify potential drugs for cardiovascular diseases caused by stress-induced genes in vascular smooth muscle cells. PeerJ 4, e2478 (2016).
Kunkel, S. D. et al. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 13, 627–638 (2011).
Malcomson, B. et al. Connectivity mapping (ssCMap) to predict A20-inducing drugs and their antiinflammatory action in cystic fibrosis. Proc. Natl Acad. Sci. USA 113, E3725–E3734 (2016).
Mirza, N., Sills, G. J., Pirmohamed, M. & Marson, A. G. Identifying new antiepileptic drugs through genomics-based drug repurposing. Hum. Mol. Genet. 26, 527–537 (2017).
Shin, E., Lee, Y. C., Kim, S. R., Kim, S. H. & Park, J. Drug signature-based finding of additional clinical use of LC28-0126 for neutrophilic bronchial asthma. Sci. Rep. 5, 17784 (2015).
Wei, G. et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell 10, 331–342 (2006).
Chiang, A. P. & Butte, A. J. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin. Pharmacol. Ther. 86, 507–510 (2009).
Iorio, F., Isacchi, A., di Bernardo, D. & Brunetti-Pierri, N. Identification of small molecules enhancing autophagic function from drug network analysis. Autophagy 6, 1204–1205 (2010).
Hegde, R. N. et al. Unravelling druggable signalling networks that control F508del-CFTR proteostasis. eLife 4, e10365 (2015).
Iorio, F. et al. A semi-supervised approach for refining transcriptional signatures of drug response and repositioning predictions. PLOS ONE 10, e0139446 (2015).
Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Wang, Z. et al. Extraction and analysis of signatures from the gene expression omnibus by the crowd. Nat. Commun. 7, 12846 (2016).
Pacini, C. et al. DvD: an R/Cytoscape pipeline for drug repurposing using public repositories of gene expression data. Bioinformatics 29, 132–134 (2013).
Zhang, S. D. & Gant, T. W. sscMap: an extensible Java application for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics 10, 236 (2009).
Oprea, T. I., Tropsha, A., Faulon, J. L. & Rintoul, M. D. Systems chemical biology. Nat. Chem. Biol. 3, 447–450 (2007).
Campillos, M., Kuhn, M., Gavin, A. C., Jensen, L. J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008).
Yang, L. & Agarwal, P. Systematic drug repositioning based on clinical side-effects. PLOS ONE 6, e28025 (2011).
Kitchen, D. B., Decornez, H., Furr, J. R. & Bajorath, J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 3, 935–949 (2004).
Dakshanamurthy, S. et al. Predicting new indications for approved drugs using a proteochemometric method. J. Med. Chem. 55, 6832–6848 (2012).
Cooke, R. M., Brown, A. J., Marshall, F. H. & Mason, J. S. Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug Discov. Today 20, 1355–1364 (2015).
Kharkar, P. S., Warrier, S. & Gaud, R. S. Reverse docking: a powerful tool for drug repositioning and drug rescue. Future Med. Chem. 6, 333–342 (2014).
Huang, H. et al. Reverse screening methods to search for the protein targets of chemopreventive compounds. Front. Chem. 6, 138 (2018).
Warren, G. L. et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 49, 5912–5931 (2006).
Pagadala, N. S., Syed, K. & Tuszynski, J. Software for molecular docking: a review. Biophys. Rev. 9, 91–102 (2017).
Sanseau, P. et al. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 30, 317–320 (2012).
Grover, M. P. et al. Novel therapeutics for coronary artery disease from genome-wide association study data. BMC Med. Genom. 8 (Suppl. 2), S1 (2015).
Wang, Z. Y. & Zhang, H. Y. Rational drug repositioning by medical genetics. Nat. Biotechnol. 31, 1080–1082 (2013).
Willyard, C. New human gene tally reignites debate. Nature 558, 354–355 (2018).
Smith, S. B., Dampier, W., Tozeren, A., Brown, J. R. & Magid-Slav, M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLOS ONE 7, e33174 (2012).
Greene, C. S. & Voight, B. F. Pathway and network-based strategies to translate genetic discoveries into effective therapies. Hum. Mol. Genet. 25, R94–R98 (2016).
Iorio, F., Saez-Rodriguez, J. & di Bernardo, D. Network based elucidation of drug response: from modulators to targets. BMC Syst. Biol. 7, 139 (2013).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
US Preventive Services Task Force. Final recommendation statement. Aspirin use to prevent cardiovascular disease and colorectal cancer: preventive medication. US Preventive Services Task Force https://www.uspreventiveservicestaskforce.org/Page/Document/RecommendationStatementFinal/aspirin-to-prevent-cardiovascular-disease-and-cancer (2017).
Cavalla, D. & Singal, C. Retrospective clinical analysis for drug rescue: for new indications or stratified patient groups. Drug Discov. Today 17, 104–109 (2012).
Jensen, P. B., Jensen, L. J. & Brunak, S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 13, 395–405 (2012).
Paik, H. et al. Repurpose terbutaline sulfate for amyotrophic lateral sclerosis using electronic medical records. Sci. Rep. 5, 8580 (2015).
Huang, Y. H. & Vakoc, C. R. A. Biomarker harvest from one thousand cancer cell lines. Cell 166, 536–537 (2016).
Weinstein, J. N. Drug discovery: cell lines battle cancer. Nature 483, 544–545 (2012).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Basu, A. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013).
Iorio, F. et al. A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016).
Seashore-Ludlow, B. et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 5, 1210–1223 (2015).
Wei, W. Q. & Denny, J. C. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 7, 41 (2015).
Chen, Z. et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol. 40, 1652–1666 (2011).
Millwood, I. Y. et al. A phenome-wide association study of a lipoprotein-associated phospholipase A2 loss-of-function variant in 90 000 Chinese adults. Int. J. Epidemiol. 45, 1588–1599 (2016).
O'Donoghue, M. L. et al. Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA 312, 1006–1015 (2014).
White, H. D. et al. Darapladib for preventing ischemic events in stable coronary heart disease. N. Engl. J. Med. 370, 1702–1711 (2014).
Eisenstein, M. Big data: the power of petabytes. Nature 527, S2–S4 (2015).
Peplow, M. The 100,000 Genomes Project. BMJ 353, i1757 (2016).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Cyranoski, D. China embraces precision medicine on a massive scale. Nature 529, 9–10 (2016).
Juric, D. et al. Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: results from the first-in-human study. J. Clin. Oncol. 36, 1291–1299 (2018).
Venot, Q. et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 558, 540–546 (2018).
Wicks, P., Vaughan, T. E., Massagli, M. P. & Heywood, J. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat. Biotechnol. 29, 411–414 (2011).
Brehmer, D. et al. Cellular targets of gefitinib. Cancer Res. 65, 379–382 (2005).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Alshareef, A. et al. The use of cellular thermal shift assay (CETSA) to study crizotinib resistance in ALK-expressing human cancers. Sci. Rep. 6, 33710 (2016).
Miettinen, T. P. & Bjorklund, M. NQO2 is a reactive oxygen species generating off-target for acetaminophen. Mol. Pharm. 11, 4395–4404 (2014).
Eyers, P. A., van den, I. P., Quinlan, R. A., Goedert, M. & Cohen, P. Use of a drug-resistant mutant of stress-activated protein kinase 2a/p38 to validate the in vivo specificity of SB 203580. FEBS Lett. 451, 191–196 (1999).
Cohen, P. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 1, 309–315 (2002).
Blagg, J. & Workman, P. Choose and use your chemical probe wisely to explore cancer biology. Cancer Cell 32, 9–25 (2017).
Hall-Jackson, C. A. et al. Paradoxical activation of Raf by a novel Raf inhibitor. Chem. Biol. 6, 559–568 (1999).
Su, F. et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366, 207–215 (2012).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Klaeger, S. et al. Chemical proteomics reveals ferrochelatase as a common off-target of kinase inhibitors. ACS Chem. Biol. 11, 1245–1254 (2016).
Troutman, S. et al. Crizotinib inhibits NF2-associated schwannoma through inhibition of focal adhesion kinase 1. Oncotarget 7, 54515–54525 (2016).
Wisniewski, D. et al. Characterization of potent inhibitors of the Bcr-Abl and the c-kit receptor tyrosine kinases. Cancer Res. 62, 4244–4255 (2002).
Blanke, C. D. et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J. Clin. Oncol. 26, 620–625 (2008).
Sloane, D. A. et al. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem. Biol. 5, 563–576 (2010).
Bago, R. et al. The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 35, 1902–1922 (2016).
Carter, T. A. et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl Acad. Sci. USA 102, 11011–11016 (2005).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Karaman, M. W. et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 (2008).
Munoz, L. Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 16, 424–440 (2017).
Xu, M. et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 22, 1101–1107 (2016).
Sun, W. et al. Rapid antimicrobial susceptibility test for identification of new therapeutics and drug combinations against multidrug-resistant bacteria. Emerg. Microbes Infect. 5, e116 (2016).
Moffat, J. G., Vincent, F., Lee, J. A., Eder, J. & Prunotto, M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 16, 531–543 (2017).
Iljin, K. et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin. Cancer Res. 15, 6070–6078 (2009).
Cousin, M. A. et al. Larval zebrafish model for FDA-approved drug repositioning for tobacco dependence treatment. PLOS ONE 9, e90467 (2014).
Kremer, S. & Jones, R. Repurposed drugs: second time lucky. Life Sciences Intellectual Property Review http://www.lifesciencesipreview.com/article/repurposed-drugs-second-time-lucky (2014).
Murteira, S., Millier, A., Ghezaiel, Z. & Lamure, M. Drug reformulations and repositioning in the pharmaceutical industry and their impact on market access: regulatory implications. J. Mark. Access Health Policy https://doi.org/10.3402/jmahp.v2.22813 (2014).
Frail, D. E. et al. Pioneering government-sponsored drug repositioning collaborations: progress and learning. Nat. Rev. Drug Discov. 14, 833–841 (2015).
Allison, M. NCATS launches drug repurposing program. Nat. Biotechnol. 30, 571–572 (2012).
Prague, J. K. et al. Neurokinin 3 receptor antagonism as a novel treatment for menopausal hot flushes: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet 389, 1809–1820 (2017).
[No authors listed.] AstraZeneca, Taiwan's NRPB launch drug discovery collaboration. Genetic Engineering & Biotechnology News https://www.genengnews.com/gen-news-highlights/astrazeneca-taiwan-39-s-nrpb-launch-drug-discovery-collaboration/81248991 (2013).
Drewry, D. H., Willson, T. M. & Zuercher, W. J. Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS). Curr. Top. Med. Chem. 14, 340–342 (2014).
Knapp, S. et al. A public-private partnership to unlock the untargeted kinome. Nat. Chem. Biol. 9, 3–6 (2013).
Billin, A. N. et al. Discovery of novel small molecules that activate satellite cell proliferation and enhance repair of damaged muscle. ACS Chem. Biol. 11, 518–529 (2016).
Elkins, J. M. et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol. 34, 95–103 (2016).
Xu, K. & Cote, T. R. Database identifies FDA-approved drugs with potential to be repurposed for treatment of orphan diseases. Brief Bioinform. 12, 341–345 (2011).
Crockett, S. D., Schectman, R., Sturmer, T. & Kappelman, M. D. Topiramate use does not reduce flares of inflammatory bowel disease. Dig. Dis. Sci. 59, 1535–1543 (2014).
Auteri, M., Zizzo, M. G. & Serio, R. GABA and GABA receptors in the gastrointestinal tract: from motility to inflammation. Pharmacol. Res. 93, 11–21 (2015).
Iorio, F. et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. USA 107, 14621–14626 (2010).
Lee, J. K., Shin, J. H., Lee, J. E. & Choi, E. J. Role of autophagy in the pathogenesis of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1852, 2517–2524 (2015).
Takata, M. et al. Fasudil, a rho kinase inhibitor, limits motor neuron loss in experimental models of amyotrophic lateral sclerosis. Br. J. Pharmacol. 170, 341–351 (2013).
Gunther, R. et al. Rho kinase inhibition with fasudil in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis-symptomatic treatment potential after disease onset. Front. Pharmacol. 8, 17 (2017).
Tonges, L. et al. Rho kinase inhibition modulates microglia activation and improves survival in a model of amyotrophic lateral sclerosis. Glia 62, 217–232 (2014).
Moschen, A. R. et al. The RANKL/OPG system is activated in inflammatory bowel disease and relates to the state of bone loss. Gut 54, 479–487 (2005).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
University of Manitoba. The effect of denosumab, the inhibitor for receptor activator of nuclear factor kappa-B ligand (RANKL), on dinitrobenzensulfonic acid (DNBS)-induced experimental model of crohn's disease. University of Manitoba MSpace https://mspace.lib.umanitoba.ca/handle/1993/32400?show=full (2017).
Gligorijevic, V., Malod-Dognin, N. & Przulj, N. Integrative methods for analyzing big data in precision medicine. Proteomics 16, 741–758 (2016).
Chen, Y., Elenee Argentinis, J. D. & Weber, G. IBM Watson: how cognitive computing can be applied to big data challenges in life sciences research. Clin. Ther. 38, 688–701 (2016).
Ritchie, M. D., Holzinger, E. R., Li, R., Pendergrass, S. A. & Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 16, 85–97 (2015).
Luo, Y. et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 573 (2017).
Napolitano, F. et al. Drug repositioning: a machine-learning approach through data integration. J. Cheminform 5, 30 (2013).
Hennequin, L. F. et al. N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J. Med. Chem. 49, 6465–6488 (2006).
Fury, M. G. et al. Phase II study of saracatinib (AZD0530) for patients with recurrent or metastatic head and neck squamous cell carcinoma (HNSCC). Anticancer Res. 31, 249–253 (2011).
Gangadhar, T. C., Clark, J. I., Karrison, T. & Gajewski, T. F. Phase II study of the Src kinase inhibitor saracatinib (AZD0530) in metastatic melanoma. Invest. New Drugs 31, 769–773 (2013).
Kaufman, A. C. et al. Fyn inhibition rescues established memory and synapse loss in Alzheimer mice. Ann. Neurol. 77, 953–971 (2015).
Nygaard, H. B. et al. A phase Ib multiple ascending dose study of the safety, tolerability, and central nervous system availability of AZD0530 (saracatinib) in Alzheimer's disease. Alzheimers Res. Ther. 7, 35 (2015).
De Felice, M., Lambert, D., Holen, I., Escott, K. J. & Andrew, D. Effects of Src-kinase inhibition in cancer-induced bone pain. Mol. Pain 12, 1744806916643725 (2016).
Tyryshkin, A., Bhattacharya, A. & Eissa, N. T. SRC kinase is a novel therapeutic target in lymphangioleiomyomatosis. Cancer Res. 74, 1996–2005 (2014).
Dickerson, J. & Sharp, F. R. Atypical antipsychotics and a Src kinase inhibitor (PP1) prevent cortical injury produced by the psychomimetic, noncompetitive NMDA receptor antagonist MK-801. Neuropsychopharmacology 31, 1420–1430 (2006).
Stearns, V. et al. Hot flushes. Lancet 360, 1851–1861 (2002).
Prague, J. K. & Dhillo, W. S. Neurokinin 3 receptor antagonism — the magic bullet for hot flushes? Climacteric 20, 505–509 (2017).
Jayasena, C. N. et al. Neurokinin B administration induces hot flushes in women. Sci. Rep. 5, 8466 (2015).
Mittelman-Smith, M. A., Williams, H., Krajewski-Hall, S. J., McMullen, N. T. & Rance, N. E. Role for kisspeptin/neurokinin B/dynorphin (KNDy) neurons in cutaneous vasodilatation and the estrogen modulation of body temperature. Proc. Natl Acad. Sci. USA 109, 19846–19851 (2012).
Rance, N. E. & Young, W. S. 3rd. Hypertrophy and increased gene expression of neurons containing neurokinin-B and substance-P messenger ribonucleic acids in the hypothalami of postmenopausal women. Endocrinology 128, 2239–2247 (1991).
Dacks, P. A., Krajewski, S. J. & Rance, N. E. Activation of neurokinin 3 receptors in the median preoptic nucleus decreases core temperature in the rat. Endocrinology 152, 4894–4905 (2011).
Crandall, C. J. et al. Association of genetic variation in the tachykinin receptor 3 locus with hot flashes and night sweats in the Women's Health Initiative Study. Menopause 24, 252–261 (2017).
Griebel, G. & Beeske, S. Is there still a future for neurokinin 3 receptor antagonists as potential drugs for the treatment of psychiatric diseases? Pharmacol. Ther. 133, 116–123 (2012).
Evangelista, S. Talnetant GlaxoSmithKline. Curr. Opin. Investig. Drugs 6, 717–721 (2005).
Fraser, G. L. et al. Clinical evaluation of the NK3 receptor antagonist fezolinetant (a.k.a. ESN364) for the treatment of menopausal hot flashes. Endocrine Society https://www.endocrine.org/meetings/endo-annual-meetings/abstract-details?ID=32994&ID=32994 (2017).
Cully, M. Deal watch: neurokinin 3 receptor antagonist revival heats up with Astellas acquisition. Nat. Rev. Drug Discov. 16, 377 (2017).
Protheroe, A., Edwards, J. C., Simmons, A., Maclennan, K. & Selby, P. Remission of inflammatory arthropathy in association with anti-CD20 therapy for non-Hodgkin's lymphoma. Rheumatology 38, 1150–1152 (1999).
Storz, U. Rituximab: how approval history is reflected by a corresponding patent filing strategy. mAbs 6, 820–837 (2014).
Brinkmann, V. et al. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9, 883–897 (2010).
Bezprozvanny, I. The rise and fall of Dimebon. Drug News Perspect. 23, 518–523 (2010).
Cudkowicz, M. E. et al. Safety and efficacy of ceftriaxone for amyotrophic lateral sclerosis: a multi-stage, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 13, 1083–1091 (2014).
Markey, K. A. et al. Assessing the efficacy and safety of an 11beta-hydroxysteroid dehydrogenase type 1 inhibitor (AZD4017) in the Idiopathic Intracranial Hypertension Drug Trial, IIH:DT: clinical methods and design for a phase II randomized controlled trial. JMIR Res. Protoc. 6, e181 (2017).
Assouline, S. et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood 114, 257–260 (2009).
Acknowledgements
This paper stems from a workshop hosted by the Medical Research Council (MRC) Centre for Drug Safety Science (CDSS; http://www.liv.ac.uk/drug-safety), University of Liverpool, in conjunction with the UK Pharmacogenetics and Stratified Medicine Network (http://www.uk-pgx-stratmed.co.uk) in November 2015 to discuss the current status of drug repurposing and evaluate various challenges faced by this field. The workshop was attended by representatives from pharmaceutical and biotechnology companies, including small-and-medium-sized enterprises focusing on drug repurposing, contract research organizations, regulatory agencies, research funding charities and academia. The content of this review has been expanded through literature search and further discussions of the authors and their research networks since the workshop.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.H. is an author on this manuscript and works for the Medicines and Healthcare Products Regulatory Agency (MHRA), UK; the opinions expressed in this review are her own and should not be attributed to the MHRA/European Medicines Agency (EMA). A.D. is a director of PharmaKure Ltd.
Related links
RELATED LINKS
Rights and permissions
About this article
Cite this article
Pushpakom, S., Iorio, F., Eyers, P. et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18, 41–58 (2019). https://doi.org/10.1038/nrd.2018.168
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2018.168