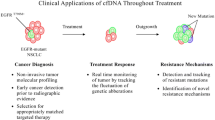
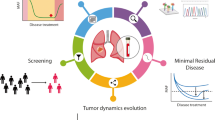
Abstract
Therapeutic advances in the treatment of lung cancer are in part due to a more complete understanding of its genomic portrait. The serial monitoring of tumor genotypes, which are instable and prone to changes under selective pressure, is becoming increasingly needed. Although tumor biopsies remain the reference standard for the diagnosis and genotyping of lung cancer, they are invasive and not always feasible. The “liquid biopsies” have the potential to overcome many of these hurdles, allowing a rapid and accurate identification of de novo and resistant genetic alterations and a real-time monitoring of treatment responses. In this review, we provide insights into new liquid diagnostic platforms and discuss the role of circulating tumor cells and circulating tumor DNA in the diagnosis and identification of resistance mutations in lung cancer.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. Cancer incidence and mortality worldwide: IARC. GLOBOCAN 2012 v1.0, CancerBase No. 11. Vol 2016. Available at: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Accessed: July 17, 2016.
Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi:10.1056/NEJMoa1102873.
Esposito A, Criscitiello C, Locatelli M, Milano M, Curigliano G. Liquid biopsies for solid tumors: understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol Ther. 2016;157:120–4. doi:10.1016/j.pharmthera.2015.11.007.
Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–82.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–8.
Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–37.
Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–20.
Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65.
Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chassé E, et al. Circulating deoxyribonucleic acid as a prognostic marker in non-small cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004;22:4157–64.
Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61:4675–8.
Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–8.
Paci M, Maramotti S, Bellesia E, Formisano D, Albertazzi L, Ricchetti T, et al. Circulating plasma DNA as diagnostic biomarker in non small-cell lung cancer. Lung Cancer. 2009;64:92–7.
Catarino R, Coelho A, Araujo A, Gomes M, Nogueira A, Lopes C, et al. Circulating DNA: diagnostic tool and predictive marker for overall survival of NSCLC patients. PLoS One. 2012;7:e38559.
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54.
Zhang R, Shao F, Wu X, Ying K. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysis. Lung Cancer. 2010;69:225–31.
Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Lo Vullo S, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med. 2009;179:69–74.
Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21:3196–203.
Marchetti A, Palma JF, Felicioni L, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol. 2015;10:1437–43.
Nygaard AD, Garm Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79:312–7.
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. North-East Japan study gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Spanish lung cancer group incollaboration with Groupe Francais de P-C and Associazione ItalianaOncologia T, erlotinib versus standard chemotherapy as first-line treatmentfor European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label,randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–22.
Perkins G, Yap TA, Pope L, Cassidy AM, Dukes JP, Riisnaes R, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One. 2012;7(11):e47020.
•• Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O'Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–705. This paper provide an evaluation of clonal genomic evolution associated with treatment response and resistance to EGFR TKIs.
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12.
•• Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–2. The study demonstrates that analysis of cf-DNA is able to detect heterogeneous mechanisms of resistance to EGFR-targeted therapies.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904.
Morrow CJ, Trapani F, Metcalf RL, Bertolini G, Hodgkinson CL, Khandelwal G, et al., Tumorigenic non-small cell lung cancer mesenchymal circulating tumour cells—a clinical case study, Ann Oncol. (2016).
Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903.
Xenidis N, Perraki M, Kafousi M, Apostolaki S, Bolonaki I, Stathopoulou A, et al. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol. 2006;24:3756–62.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904.
Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31(2):427–41.
Di Carlo D, Edd JF, Irimia D, Tompkins RG, Toner M. Equilibrium separation and filtration of particles using differential inertial focusing. Anal Chem. 2008;80(6):2204–11.
Huang T, Jia CP, Jun Y, Sun WJ, Wang WT, Zhang HL, et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron. 2014;51:213–8.
Hosokawa M, Yoshikawa T, Negishi R, Yoshino T, Koh Y, Kenmotsu H, et al. Microcavity array system for size-based enrichment of circulating tumor cells from the blood of patients with small-cell lung cancer. Anal Chem. 2013;85(12):5692–8.
Fan X, Jia C, Yang J, Li G, Mao H, Jin Q, Zhao J. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens Bioelectron. 2015;71:380–6.
Tanaka F, Yoneda K, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res. 2009;15:6980–6.
Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, et al. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014;9:e111597.
Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg. 2015;99(6):1899–905.
Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5(180):180ra48.
Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–63.
Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung cancer cells. N Engl JMed. 2008;359:366–77.
Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9(8):e103883.
Breitenbuecher F, Hoffarth S, Worm K, Cortes-Incio D, Gauler TC, Köhler J, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014;9:e85350.
Ilie M, Long E, Butori C, Hofman V, Coelle C, Mauro V, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol. 2012;23:2907–13.
Pailler E, Adam J, Barthelemy A, Oulhen M, Auger N, Valent A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013;31:2273–81.
•• Pailler E, Auger N, Lindsay CR, Vielh P, Islas-Morris-Hernandez A, Borget I, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol (2015) 26(7):1408–1415. ROS1 rearrangement was investigated in NSCLC CTCs and compared to tumor biopsy samples.
Klein CA, Seidl S, Petat-Dutter K, Offner S, Geigl JB, Schmidt-Kittler O, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotechnol. 2002;20:387–92.
Tsimberidou AM, Wen S, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–83.
Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci TranslMed. 2011;3:75ra26.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Angela Esposito, Carmen Criscitiello, Dario Trapani, and Giuseppe Curigliano declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Lung Cancer
Rights and permissions
About this article
Cite this article
Esposito, A., Criscitiello, C., Trapani, D. et al. The Emerging Role of “Liquid Biopsies,” Circulating Tumor Cells, and Circulating Cell-Free Tumor DNA in Lung Cancer Diagnosis and Identification of Resistance Mutations. Curr Oncol Rep 19, 1 (2017). https://doi.org/10.1007/s11912-017-0564-y
Published:
DOI: https://doi.org/10.1007/s11912-017-0564-y