Abstract
Lynch syndrome is a cancer-predisposing syndrome inherited in an autosomal-dominant manner, wherein colon cancer and endometrial cancer develop frequently in the family, it results from a loss-of-function mutation in one of four different genes (MLH1, MSH2, MSH6, and PMS2) encoding mismatch repair proteins. Being located immediately upstream of the MSH2 gene, EPCAM abnormalities can affect MSH2 and cause Lynch syndrome. Mismatch repair proteins are involved in repairing of incorrect pairing (point mutations and deletion/insertion of simple repetitive sequences, so-called microsatellites) that can arise during DNA replication. MSH2 forms heterodimers with MSH6 or MSH3 (MutSα, MutSβ, respectively) and is involved in mismatch-pair recognition and initiation of repair. MLH1 forms a complex with PMS2, and functions as an endonuclease. If the mismatch repair system is thoroughly working, genome integrity is maintained completely. Lynch syndrome is a state of mismatch repair deficiency due to a monoallelic abnormality of any mismatch repair genes. The phenotype indicating the mismatch repair deficiency can be frequently shown as a microsatellite instability in tumors. Children with germline biallelic mismatch repair gene abnormalities were reported to develop conditions such as gastrointestinal polyposis, colorectal cancer, brain cancer, leukemia, etc., and so on, demonstrating the need to respond with new concepts in genetic counseling. In promoting cancer genome medicine in a new era, such as by utilizing immune checkpoints, it is important to understand the genetic and genomic molecular background, including the status of mismatch repair deficiency.






Similar content being viewed by others
Change history
31 July 2019
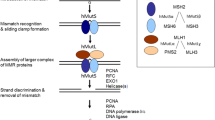
In the original publication, part d of Figure 2 was mistakenly not included.
Abbreviations
- MMR:
-
Mismatch repair
- MSI:
-
Microsatellite instability
- PCNA:
-
Proliferating cellular nuclear antigen
- RFC:
-
Replication factor
- CTE:
-
Congenital tufting enteropathy
- CMMR-D:
-
Constitutional mismatch repair deficiency
- CNS:
-
Central nervous system
- IHC:
-
Immunohistochemical staining
- MLPA:
-
Multiple ligation-dependent probe amplification
- CTLA-4:
-
Cytotoxic T-lymphocyte-associated protein 4
- PD-1:
-
Programmed cell death protein 1
- TMB:
-
Tumor mutational burden
- ICI:
-
Immune checkpoint inhibitor
References
Vogelstein B, Fearon ER, Hamilton SR et al (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319:525–532
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61:759–767
Bodmer W, Bishop T, Karran P (1994) Genetic steps in colorectal cancer. Nature Genet 6:217–219
Kinzler KW, Vogelstein B (1996) Lessons from hereditary colorectal cancer. Cell 87:159–170
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Tamura K, Utsunomiya J, Iwama T et al (2004) Mechanism of carcinogenesis in familial tumors. Int J Clin Oncol 9:232–245
Lynch HT, de la Chapelle A (2003) Hereditary colorectal cancer. N Engl J Med 348:919–932
Holliday R (1964) A mechanism for gene conversion in fungi. Genet Res 5:282–304
Modrich P (2016) Mechanisms in E. coli and mismatch repair. Angew Chem Int Ed Engl 55:8490–8501
Nevers P, Spats HC (1975) Escherichia coli mutants uvr D and uvr E deficient in gene conversion of lambda-heteroduplexes. Mol Gen Genet 139:233–243
Rydberg B (1978) Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res 52:11–24
Glickman BW, Radman M (1980) Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci USA 77:1063–1067
Lauhe RS, Su SS, Morich P (1987) Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci USA 84:1482–1486
Su SS, Morrich P (1986) Escherichia coli mutS-encoded protein binds to Mismatched DNA base pairs. Proc Natl Acad Sci USA 83:5057–5061
Meselson M (1988) Methyl-directed repair of DNA mismatches. In: Low KB (ed) Recombination of the genetic material. Academic Press, San Diego, pp 91–113
Modrich P (1989) Methyl-directed DNA mismatch correction. J Biol Chem 264:6597–6600
Grilley M, Holmes J, Yashar B et al (1990) Mechanisms of DNA-mismatch correction. Mutat Res 236:253–267
Modrich P (1991) Mechanisms and biological effects of mismatch repair. Annu Rev Genet 25:229–253
Ligtenberg MJL, Kuiper RP, Chan TL et al (2009) Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3-prime exons of TACSTD1. Nature Genet 41:112–117
Fishel R, Lescoe MK, Rao MRS et al (1993) The human mutator gene homolog MSH2 and its association. Cell 75:1027–1038
Leach FS, Nicolaides NC, Papadopoulos N et al (1993) Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215–1225
Kolodner RD, Hall NR, Lipford J et al (1994) Structure of the human MSH2 locus and analysis of two Muir-Torre kindreds for msh2 mutations. Genomics 24:516–526
Fishel R, Ewel A, Lee S et al (1993) Binding of mismatched microsatellite DNA sequences by the human MSH2 protein. Science 266:1403–1405
Papadopoulos N, Nicolaides NC, Wei Y-F et al (1994) Mutation of a mutL homolog in hereditary colon cancer. Science 263:1625–1629
Bronner CE, Baker SM, Morrison PT et al (1994) Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368:258–261
Han H-J, Maruyama M, Baba S et al (1995) Genomic structure of human mismatch repair gene, hMLH1, and its mutation analysis in patients with hereditary non-polyposis colorectal cancer (HNPCC). Hum Molec Genet 4:237–242
Drummond JT, Li G-M, Longley MJ et al (1995) Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268:1909–1912
Palombo F, Gallinari P, Iaccarino I et al (1995) GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science 268:1912–1914
Miyaki M, Konishi M, Tanaka K et al (1997) Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nature Genet 17:271–272
Akiyama Y, Sato H, Yamada T et al (1997) Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res 57:3920–3923
Nicolaides NC, Papadopoulos N, Liu B et al (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371:75–80
De Vos M, Hayward BE, Picton S et al (2004) Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet 74:954–964
Ban C, Juno M, Yang W (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97:85–97
Tran PT, Liskay RM (2000) Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol Cell Biol 20:6390–6398
Räschle M, Dufner P, Marra G et al (2002) Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem 277:21810–21820
Guerrette S, Acharya S, Fishel R (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 274:6336–6341
Kondo E, Horii A, Fukushige S (2001) The interaction domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res 29:1695–1708
Reenan RA, Kolodner RD (1992) Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132:963–973
Li GM, Modrich P (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA 92:1950–1954
Johnson RE, Kovvali GK, Guzder SN et al (1996) Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem 271:27987–27990
Umar A, Buermeyer AB, Simon JA et al (1996) Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65–73
Tishkoff DX, Boerger AL, Bertrand P et al (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA 94:7487–7492
Longley MJ, Pierce AJ, Modrich P (1997) DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem 272:10917–10921
Schmutte C, Marinescu RC, Sadoff MM et al (1998) Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res 58:4537–4542
Tishkoff DX, Amin NS, Viars CS et al (1998) Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 58:5027–5031
Lin YL, Shivji MK, Chen C et al (1998) The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem 273:1453–1461
Gu L, Hong Y, McCulloch S et al (1998) ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res 26:1173–1178
Zhang Y, Yuan F, Presnell SR et al (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122:693–705
Genschel J, Littman SJ, Drummond JT et al (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem 273(31):19895–19901
Iyer RR, Pluciennik A, Genschel J et al (2010) MutLalpha and proliferating cell nuclear antigen share binding sites on MutSbeta. J Biol Chem 285(15):11730–11739
Plotz G, Raedle J, Brieger A et al (2003) N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha. Nucleic Acids Res 31(12):3217–3226
Dahal BK, Kadyrova LY, Delfino KR et al (2017) Involvement of DNA mismatch repair in the maintenance of heterochromatic DNA stability in Saccharomyces cerevisiae. PLoS Genet 13(10):e1007074
Villahermosa D, Christensen O, Knapp K et al (2017) Schizosaccharomyces pombe MutSα and MutLα maintain stability of tetra-nucleotide repeats and Msh3 of hepta-nucleotide repeats. G3 (Bethesda) 7(5):1463–1473
Prolla TA, Baker SM, Harris AC et al (1998) Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet 18(3):276–279
Jäger AC, Rasmussen M, Bisgaard HC et al (2001) HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene 20(27):3590–3595
Cannavo E, Marra G, Sabates-Bellver J et al (2005) Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res 65(23):10759–10766
Kadyrov FA, Dzantiev L, Constantin N et al (2006) Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126(2):297–308
Peng M, Litman R, Xie J, Sharma S, Brosh RM Jr, Cantor SB (2007) The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J 26(13):3238–3249
Pluciennik A, Dzantiev L, Iyer RR et al (2010) PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA 107(37):16066–16071
Kunkel TA, Erie DA (2005) DNA mismatch repair. Annu Rev Biochem 74:681–710
Li GM (2008) Mechanisms and functions of DNA mismatch repair. Cell Res 18(1):85–98
Kunkel TA, Erie DA (2015) Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet 49:291–313
Marti TM, Kunz C, Fleck O (2002) DNA mismatch repair and mutation avoidance pathways. J Cell Physiol 191(1):28–41
Friedberg EC (2003) DNA damage and repair. Nature 421(6921):436–440
Martin SA, Lord CJ, Ashworth A (2010) Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res 16(21):5107–5113
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138(6):2073–2087
Fishel R (2015) Mismatch repair. J Biol Chem 290(44):26395–26403
Groothuizen FS, Sixma TK (2016) The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst) 38:14–23
Hingorani MM (2016) Mismatch binding, ADP-ATP exchange and intramolecular signaling during mismatch repair. DNA Repair (Amst) 38:24–31
Kadyrova LY, Kadyrov FA (2016) Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Repair (Amst) 38:42–49
Peltomäki P (2016) Update on Lynch syndrome genomics. Fam Cancer 15:385–393
Liu D, Keijzers G, Rasmussen LJ (2017) DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res 773:174–187
Lee JB, Cho WK, Park J et al (2014) Single-molecule views of MutS on mismatched DNA. DNA Repair 20:82–93
Plotz G, Raedle J, Brieger A et al (2002) hMutSalpha forms an ATP-dependent complex with hMutLalpha and hMutLbeta on DNA. Neucleic Acids Res 30(3):711–718
Plotz G, Piiper A, Wormek M et al (2006) Analysis of the human MutLalpha.MutSalpha. Biochem Biophys Res Commun. 340(3):852–859
Friedhoff P, Li P, Gotthardt J (2016) Protein-protein interactions in DNA mismatch repair. DNA Repair 38:50–57
Jeon Y, Kim D, Martin-Lopez JV et al (2016) Dynamic control of strand excision during human DNA mismatch repair. Proc Natl Acad Sci USA 113(12):3281–3286
Fishel R (1998) Mismatch repair, molecular switches, and signal transduction. Genes Dev 12(14):2096–2101
Ban C, Junop M, Yang W (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97(1):85–97
Spampinato C, Modrich P (2000) The MutL ATPase is required for mismatch repair. J Biol Chem 275(13):9863–9869
Lamers MH, Winterwerp HH, Sixma TK (2003) The alternating ATPase domains of MutS control DNA mismatch repair. ENBO J 22(3):746–756
Kolodner RD, Marsischky GT (1999) Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9(1):89–96
Peltomäki P (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet 10(7):735–740
Bellacosa A (2001) Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ 8(11):1076–1092
Scmidt MHM, Pearson CE (2016) Disease associated repeat instability and mismatch repair. DNA Repair 38:117–126
Campregher C, Schmid G, Ferk F et al (2012) MSH3-deficiency initiates EMAST without oncogenic transformation of human colon epithelialcells. PLoS One 7(11):e50541. https://doi.org/10.1371/journal.pone.0050541(Epub 2012 Nov 27)
Tseng-Rogenski SS, Chung H, Wilk M et al (2012) Oxidative stress induces nuclear-to-cytosol shift of hMSH3, a potential mechanism for EMAST in colorectal cancer cells. PLoS One 7(11):e50616. https://doi.org/10.1371/journal.pone.0050616(Epub 2012 Nov 30)
Watson MMC, Berg M, Søreide K (2014) Prevalence and implications of elevated microsatellite alterations at selected tetranucleotides in cancer. Br J Cancer 111(5):823–827
Carethers JM, Koi M, Tseng-Rogenski SS (2015) EMAST is a form of microsatellite instability that is initiated by inflammation and modulates colorectal cancer progression. Genes 6(2):185–205
Venderbosch S, van Lent-van Vliet S, de Haan AF et al (2015) EMAST is associated with a poor prognosis in microsatellite instable metastatic colorectal cancer. PLoS One 10(4):e0124538. https://doi.org/10.1371/journal.pone.0124538.eCollection
Dollé E, Theise ND, Schmelzer E et al (2015) EpCAM and the biology of hepatic stem/progenitor cells. Am J Physiol Gastrointest Liver Physiol 308:G233–G250
Huang L, Yang Y, Yang F et al (2018) Functions of EpCAM in physiological processes and diseases. Int J Mol Med 42(4):1771–1785
Kovacs ME, Papp J, Szentirmay Z et al (2009) Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat 30(2):197–203
Sivagnanam M, Mueller JL, Lee H et al (2008) Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterol 135(2):429–437
Reifen RM, Cutz E, Griffiths AM et al (1994) Tufting enteropathy: a newly recognized clinicopathological entity associated with refractory diarrhea in infants. J Pediatr Gastroenterol Nutr 18(3):379–385
Goulet O, Salomon J, Ruemmele F et al (2007) Intestinal epithelial dysplasia (tufting enteropathy). Orphanet J Rare Dis 2(1):20
Pathak SJ, Mueller JL, Okamoto K et al (2019) EPCAM mutation update: variants associated with congenital tufting enteropathy and Lynch syndrome. Hum Mutat 40(2):142–161
Wimmer K, Kratz CP, Vasen HFA et al (2014) Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J Med Genet 51(6):355–365
Ricciardone MD, Ozçelik T, Cevher B et al (1999) Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res 59(2):290–293
Wang Q, Lasset C, Desseigne F et al (1999) Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res 59(2):294–297
Turcot J, Despres JP, St Pierre F et al (1959) Malignant tumors of the central nervous system associated with familial polyposis of the colon: report of two cases. Dis Colon Rectum 2:465–468
Hamilton SR, Liu B, Parsons RE et al (1995) The molecular basis of Turcot’s syndrome. N Engl J Med 332(13):839–847
Lavoine N, Colas C, Muleris M et al (2015) Constitutional mismatch repair deficiency syndrome: clinical description in a French cohort. J Med Genet 52(11):770–778
Turcot J, Despres JP, St Pierre F (1959) Malignant tumors of the central nervous system associated with familial polyposis of the colon: report of two cases. Dis Colon Rectum 2(5):465–468
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group (2009) Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 11(1):35–41
Giardiello FM, Allen JI, Axilbund JE et al (2014) Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multisociety Task Force on colorectal cancer. Am J Gastroenterol 109(8):1159–1179
Syngal S, Brand RE, Church JM et al (2015) ACG clinical guideline: genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 110(2):223–262
Boland CR, Thibodeau SN, Hamilton SR et al (1998) A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58(22):5248–5257
Ionov Y, Peinado MA, Malkhosyan S et al (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363(6429):558–561
Peltomäki P, Aaltonen LA, Sistonen P et al (1993) Genetic mapping of a locus predisposing to human colorectal cancer. Nature 260:810–812
Thibodeau SN, Bren G, Schaid D (1993) Microsatellite instability in cancer of the proximal colon. Nature 260(5109):816–819
Rodriguez-Bigas MA, Boland CR, Hamilton SR et al (1997) A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89(23):1758–1762
Leach FS, Polyak K, Burrell M et al (1996) Expression of the human mismatch repair gene hMSH2 in normal and neoplastic tissues. Cancer Res 56(2):235–240
Thibodeau SN, French AJ, Roche PC et al (1996) Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 56(21):4836–4840
Hendriks Y, Franken P, Dierssen JW et al (2003) Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 162(2):469–477
de Jong AE, van Puijenbroek M, Hendriks Y et al (2004) Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer res 10(39):972–980
Lipkin SM, Wang V, Jacoby R et al (2000) MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet 24(1):27–35
Rigau V, Sebbagh N, Olschwang S et al (2003) Microsatellite instability in colorectal carcinoma. The comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 [correction of hMLH6] immunostaining. Arch Pathol Lab Med 127(6):694–700
Hampel H, Frankel WL, Martin E et al (2005) Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352(18):1851–1860
Hendriks YMC, de Jong AE, Morreau H et al (2006) Dianostic approach and management of Lynch syndrome (hereditary nonpolyposis colorectal carcinoma): a guide for clinicians. CA Cancer J Clin 56(4):213–225
Snowsill T, Coelho H, Huxley N et al (2017) Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation. Health Technol Assess 21(51):1–238
Jin M, Hampel H, Zhou X et al (2013) BRAF V600E mutation analysis simplifies the testing algorithm for Lynch Syndrome. Am J Clin Pathol 140(2):177–183
Lagerstedt-Robinson K, Rohlin A, Aravidis C et al (2016) Mismatch repair gene mutation spectrum in the Swedish Lynch syndrome population. Oncol Rep 36(5):2823–2835
Lorans M, Dow E, Macrae FA et al (2018) Update on hereditary colorectal cancer: improving the clinical utility of multigene panel testing. Clin Colorectal Cancer 17(2):e293–e305. https://doi.org/10.1016/j.clcc.2018.01.001(Epub 2018 Jan 11)
Gallego CJ, Shirts BH, Bennette CS et al (2015) Next-generations sequencing panels for the diagnosis of colorectal cancer and polyposis syndromes: a cost-effectiveness analysis. J Clin Oncol 33(18):2084–2091
Espenschied CR, LaDuca H, Li S et al (2017) Multigene panel testing provides a new perspective on Lynch syndrome. J Clin Oncol 35(22):2568–2575
Yurgelun MB, Kulke MH, Fuchs CS et al (2017) Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol 35(10):1086–1095
Thompson BA, Spurdle AB, Plazzer JP et al (2014) Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 46(12):107–115
Roberts ME, Jackson SA, Susswein LR et al (2018) MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet Med 20(10):1167–1174
Plazzer JP, Sijmons RH, Woods MO et al (2013) The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer 12(2):175–180
Leach DR, Krummel MF, Allison JP (1996) Enhancement of antitumor immunity by CTLA-4 blockade. Science 271(5256):1734–1736
Ishida Y, Agata Y, Shibahara K, Honjo T (1992) Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 11(11):3887–3895
Tivol EA, Borriello F, Schweitzer AN et al (1995) Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 3(5):541–547
Nishimura H, Nose M, Hiai H et al (1999) Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11(2):141–151
Topalian SL, Hodi S, Brahmer JR et al (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366(26):2443–2454
Le DT, Uram JN, Wang H et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372(26):2509–2520
Snyder A, Makarov V, Merghoub T et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199
Rizvi NA, Hellmann MD, Snyder A et al (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1blockade in non-small cell lung cancer. Science 348(6230):124–128
Yarchoan M, Hopkins A, Jaffee EM et al (2018) Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 377(25):2500–2501
Charmers ZR, Connelly CF, Fabrizio D et al (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9(1):34. https://doi.org/10.1186/s13073-017-0424-2
Dudley JC, Lin MT, Le DT et al (2016) Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res 22(4):813–820
Le DT, Durham JN, Smith KN et al (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413
Acknowledgements
This work was supported in part by a grant from the Japanese Ministry of Education, Science, Sports and Culture of Japan (19K07763).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: In the original publication, Fig. 2d has not been included. Complete Figure 2 is replaced. “MLH1, MSH2, MSH6, PMS2” in legend of figure 2 are made roman. The section, “Effectiveness of immune check point blockades and a hypermutable state (high tumor mutation burden)” is revised to “Effectiveness of immune check point blockades and a hypermutable state (high tumor mutational burden)”.
About this article
Cite this article
Tamura, K., Kaneda, M., Futagawa, M. et al. Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome. Int J Clin Oncol 24, 999–1011 (2019). https://doi.org/10.1007/s10147-019-01494-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-019-01494-y